Polymer Physics
Kinetics of
interior loop formation in semiflexible chains (JCP '06)
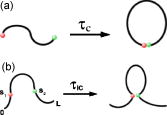
Loop
formation
between monomers in the interior of semiflexible chains describes
elementary
events in biomolecular folding and DNA bending. We calculate
analytically
the interior distance distribution function for semiflexible chains
using
a mean field approach. Using the potential of mean
force
derived from the distance distribution function we
present a simple
expression for the kinetics of interior
looping by adopting Kramers
theory. For the parameters,
that are appropriate for DNA, the
theoretical predictions
in comparison with the case are in excellent
agreement with
explicit Brownian dynamics simulations of wormlike chain (WLC)
model.
The interior looping times (
 IC
IC) can be greatly altered
in
the cases when the stiffness of the loop differs
from that
of the dangling ends. If the dangling end
is stiffer than
the loop then
 IC
IC
increases for the
case of the WLC with uniform persistence
length. In contrast,
attachment of flexible dangling ends
enhances rate of interior loop
formation. The theory also
shows that if the monomers are
charged and interact via
screened Coulomb potential then both the
cyclization (
 c
c)
and interior looping (
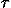 IC
IC) times greatly increase at
low
ionic concentration. Because both
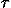 c
c and
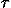 IC
IC are determined
essentially by the effective persistence
length [
l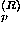
] we
computed
l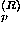
by
varying the range of the repulsive interaction between the
monomers.
For short range interactions
l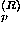
nearly
coincides with the
bare persistence length which is
determined largely by the backbone
chain connectivity. This
finding rationalizes the efficacy of describing a
number of
experimental observations (response of biopolymers to force and
cyclization
kinetics) in biomolecules using WLC model with an effective
persistence
length.
Stretching homopolymers
(Macromolecules '07)
Force-induced stretching of polymers is important in a variety of
contexts. We have used theory
and simulations to describe the response of homopolymers, with
N
monomers, to an external force (
f ) in good
and poor solvents. In good solvents and for sufficiently large
N
we show, in accord with scaling predictions, that
the mean extension along the
f axis <
Z> ~
f
for small
f and <
Z> ~
f 2/3
(the Pincus regime) for intermediate values
of
f. The theoretical predictions for <
Z> as a
function of
f are in excellent agreement with simulations for
N
= 100
and 1600. However, even with
N = 1600, the expected Pincus
regime is not observed due to the breakdown of
the assumptions in the blob picture for finite
N. We predict
the Pincus scaling in a good solvent will be observed
for
N 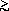
10
5. The force-dependent
structure factors for a polymer in a poor solvent show that there is a
hierarchy
of structures, depending on the nature of the solvent. For a weakly
hydrophobic polymer, various structures
(ideal conformations, self-avoiding chains, globules, and rods) emerge
on distinct length scales as
f is varied. A
strongly hydrophobic polymer remains globular as long as
f is
less than a critical value
fc. Above
fc,
an abrupt
first-order transition to a rodlike structure occurs. Our predictions
can be tested using single molecule experiments.