|

|
|
 

  
|
|
Deciphering
the energy landscape
of biomolecules
Hidden Complexity in
the Isomerization
Dynamics of Holliday
Junctions (Nature
Chem. '13)
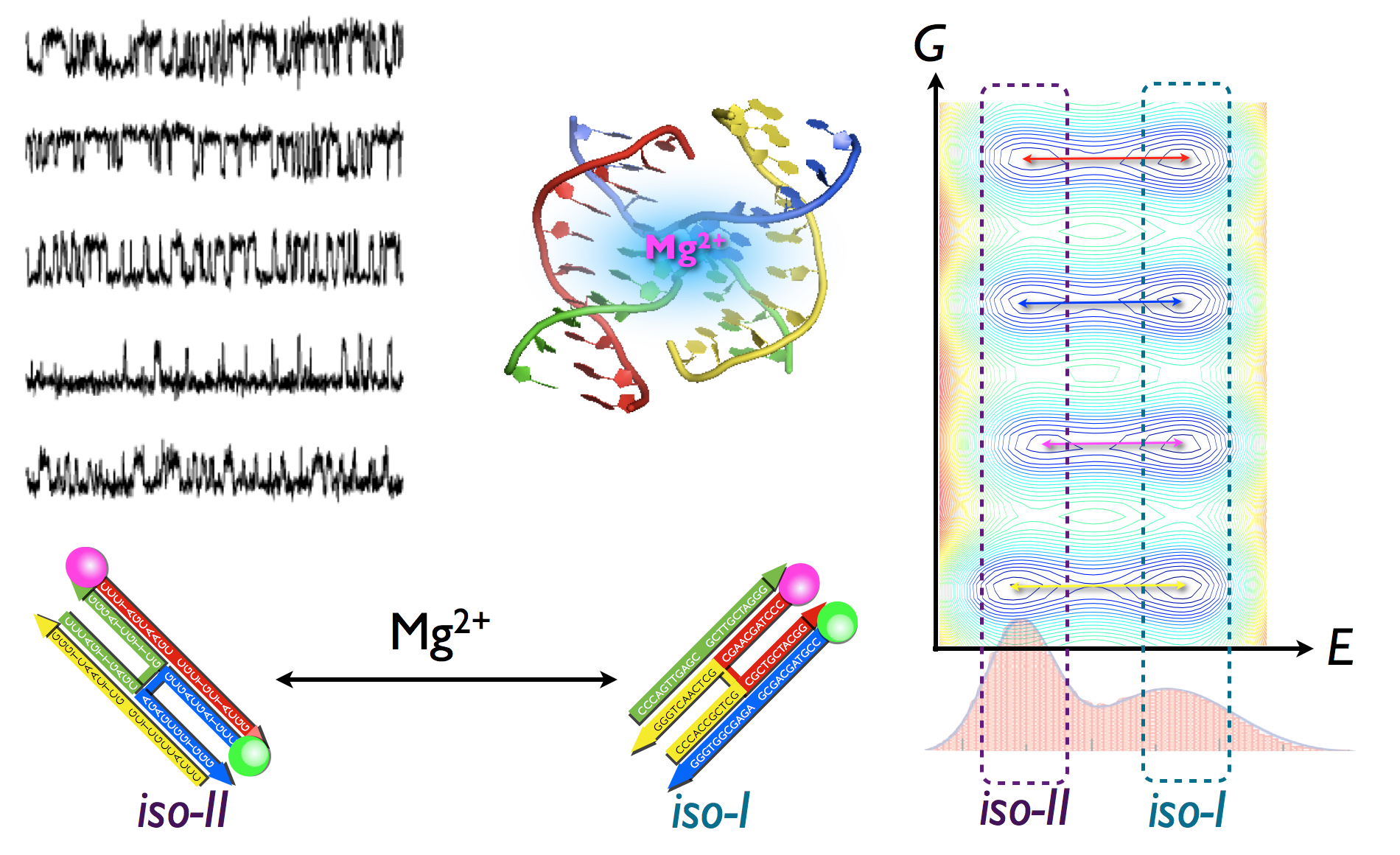
A plausible
consequence of
the rugged
folding energy
landscapes
inherent to
biomolecules
is that there
may be more
than one
functionally
competent
folded state.
Indeed,
molecule-to-molecule
variations in
the folding
dynamics of
enzymes and
ribozymes have
recently been
identified in
single-molecule
experiments,
but without
systematic
quantification
or an
understanding
of their
structural
origin. Here,
using concepts
from glass
physics and
complementary
clustering
analysis, we
provide a
quantitative
method to
analyse
single-molecule
fluorescence
resonance
energy
transfer
(smFRET) data,
thereby
probing the
isomerization
dynamics of
Holliday
junctions,
which display
such
heterogeneous
dynamics over
a long
observation
time (Tobs
≈ 40 s). We
show that the
ergodicity of
Holliday
junction
dynamics is
effectively
broken and
that their
conformational
space is
partitioned
into a folding
network of
kinetically
disconnected
clusters.
Theory
suggests that
the persistent
heterogeneity
of Holliday
junction
dynamics is a
consequence of
internal
multiloops
with varying
sizes and
flexibilities
frozen by Mg2+
ions. An
annealing
experiment
using Mg2+
pulses lends
support to
this idea by
explicitly
showing that
interconversions
between
trajectories
with different
patterns can
be induced.
Can energy landscape
roughness of
proteins and RNA be
measured by using
mechanical unfolding
experiments? (PNAS
'03)
By
considering temperature
effects on the mechanical
unfolding rates
of proteins and
RNA, whose energy
landscape is rugged,
the question posed
in the title is answered
in the affirmative. Adopting
a
theory by Zwanzig
[Zwanzig, R. (1988) Proc.
Natl. Acad.
Sci. USA 85,
2029-2030], 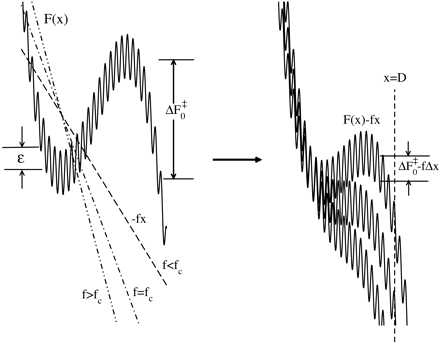 we show
that, because of
roughness characterized
by an energy scale we show
that, because of
roughness characterized
by an energy scale  , the
unfolding rate at
constant force
is retarded. Similarly, in
nonequilibrium
experiments done
at constant loading rates,
the most
probable unfolding force
increases because of
energy
landscape roughness. The
effects are dramatic
at low
temperatures. Our analysis
suggests that,
by using
temperature as a variable
in mechanical unfolding
experiments
of proteins and RNA, the
ruggedness energy scale , the
unfolding rate at
constant force
is retarded. Similarly, in
nonequilibrium
experiments done
at constant loading rates,
the most
probable unfolding force
increases because of
energy
landscape roughness. The
effects are dramatic
at low
temperatures. Our analysis
suggests that,
by using
temperature as a variable
in mechanical unfolding
experiments
of proteins and RNA, the
ruggedness energy scale
 , can be
directly measured. , can be
directly measured.
|
|
|
Contact
Information :
Changbong Hyeon,
Professor, School of Computational
Sciences, Korea Institute for Advanced
Study
, Seoul 02455, Republic of Korea
+82-2-958-3810
(tel)  |
|
© 2010 KIAS Theoretical and
Computational Biophysics
Group |
|
|