|

|
|
 

  
|
Molecular Motors
Internal strain regulates the nucleotide
binding site of the kinesin leading head (PNAS '07)
In the
presence of ATP, kinesin proceeds along
the protofilament of
microtubule by alternated binding of
two motor domains on the tubulin
binding
sites. Because the processivity of kinesin is
much higher than other motor proteins, it has been speculated that
there exists a mechanism for allosteric
regulation
between the two monomers. Recent experiments suggest that
ATP
binding to the leading head (L) domain in kinesin rearward strain built on the
neck-linker. We
test this hypothesis by explicitly modeling a C -based kinesin structure
whose motor domains are
bound on the tubulin binding sites.
The equilibrium structures
of kinesin
on the microtubule show
disordered and ordered neck-linker configurations for the L
and
trailing head, respectively. The comparison of
the structures
between the two heads shows that several native contacts
present at
the nucleotide binding site in the L are less intact than
those in
the binding site of the rear head. The
network of native contacts
obtained from this comparison provides the internal tension
propagation pathway, which leads to the disruption
of the nucleotide
binding site in the L. Also, using an argument based on
polymer
theory, we estimate the internal tension built on the
neck-linker to be f -based kinesin structure
whose motor domains are
bound on the tubulin binding sites.
The equilibrium structures
of kinesin
on the microtubule show
disordered and ordered neck-linker configurations for the L
and
trailing head, respectively. The comparison of
the structures
between the two heads shows that several native contacts
present at
the nucleotide binding site in the L are less intact than
those in
the binding site of the rear head. The
network of native contacts
obtained from this comparison provides the internal tension
propagation pathway, which leads to the disruption
of the nucleotide
binding site in the L. Also, using an argument based on
polymer
theory, we estimate the internal tension built on the
neck-linker to be f 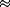 12-15 pN. Both of these
conclusions support the experimental
hypothesis. 12-15 pN. Both of these
conclusions support the experimental
hypothesis.
Mechanical control of the directional stepping dynamics of the kinesin
motor (PNAS '07)
Among
the multiple steps constituting the kinesin
mechanochemical
cycle, one of the most
interesting events is observed when kinesins
move
an 8-nm step from one microtubule (MT)-binding site to another.
The
stepping motion that occurs within a relatively short time
scale (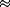 100
microsec) is, however, beyond the resolution of current
experiments.
Therefore, a basic understanding to the real-time dynamics
within
the 8-nm step is still lacking. For instance, the rate of
power
stroke (or conformational change) that leads to the
undocked-to-docked transition of neck-linker is not known,
and the
existence of a substep during the 8-nm
step
still remains a controversial issue in the kinesin
community. By using explicit structures of the kinesin dimer and
the MT consisting of 13 protofilaments,
we study the stepping dynamics with
varying rates of power stroke (kp).
We estimate that k 100
microsec) is, however, beyond the resolution of current
experiments.
Therefore, a basic understanding to the real-time dynamics
within
the 8-nm step is still lacking. For instance, the rate of
power
stroke (or conformational change) that leads to the
undocked-to-docked transition of neck-linker is not known,
and the
existence of a substep during the 8-nm
step
still remains a controversial issue in the kinesin
community. By using explicit structures of the kinesin dimer and
the MT consisting of 13 protofilaments,
we study the stepping dynamics with
varying rates of power stroke (kp).
We estimate that k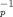 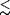 20 microsec
to avoid a substep in an
averaged time
trace. For a slow power stroke with k 20 microsec
to avoid a substep in an
averaged time
trace. For a slow power stroke with k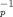 > 20 microsec, the averaged time trace shows a substep
that implies the existence of a transient intermediate,
which is
reminiscent of a recent single-molecule experiment at high
resolution. We identify the intermediate as a conformation
in which
the tethered head is trapped in the sideway binding site of
the
neighboring protofilament. We also find
a
partial unfolding (cracking) of the binding motifs occurring at
the
transition state ensemble along the pathways before binding between
the kinesin and MT.
> 20 microsec, the averaged time trace shows a substep
that implies the existence of a transient intermediate,
which is
reminiscent of a recent single-molecule experiment at high
resolution. We identify the intermediate as a conformation
in which
the tethered head is trapped in the sideway binding site of
the
neighboring protofilament. We also find
a
partial unfolding (cracking) of the binding motifs occurring at
the
transition state ensemble along the pathways before binding between
the kinesin and MT.
Dynamics
of allosteric transitions in GroEL (PNAS '06)
The chaperonin GroEL-GroES, a machine that helps
proteins to fold,
cycles through a number of allosteric states, the T state,
with high affinity for substrate
proteins, the ATP-bound R state, and the R''
(GroEL-ADP-GroES) complex. Here, we
use a self-organized polymer model for
the GroEL allosteric states and a general structure-based
technique to simulate the dynamics of allosteric
transitions in two subunits of GroEL and the heptamer. The T
 R
transition, in which the apical domains undergo
counterclockwise motion, is mediated by a multiple salt-bridge switch
mechanism, in which a series of salt-bridges break and form.
The initial event in the R R
transition, in which the apical domains undergo
counterclockwise motion, is mediated by a multiple salt-bridge switch
mechanism, in which a series of salt-bridges break and form.
The initial event in the R  R'' transition, during which GroEL
rotates clockwise, involves a spectacular outside-in movement of
helices K and L that results in K80-D359 salt-bridge formation. In
both the transitions there is considerable heterogeneity in
the transition pathways. The transition state ensembles (TSEs) connecting
the T, R, and R'' states are broad with the TSE
for the T R'' transition, during which GroEL
rotates clockwise, involves a spectacular outside-in movement of
helices K and L that results in K80-D359 salt-bridge formation. In
both the transitions there is considerable heterogeneity in
the transition pathways. The transition state ensembles (TSEs) connecting
the T, R, and R'' states are broad with the TSE
for the T  R transition being more plastic than the
R
R transition being more plastic than the
R  R'' TSE. R'' TSE.
|
|
|
Contact Information :
Changbong Hyeon,
Professor, School of Computational Sciences, Korea Institute for Advanced Study, Seoul 02455, Republic of Korea
+82-2-958-3810 (tel)
 |
|
© 2010 KIAS Theoretical and Computational Biophysics
Group |
|
|