|

|
|
 

  
|
Stretching Biomolecules
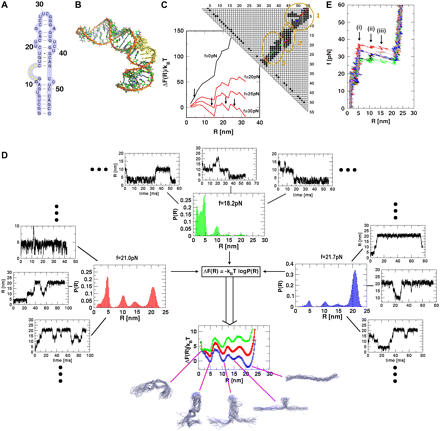
Mechanical Unfolding of RNA hairpins (PNAS '05)
Mechanical
unfolding trajectories, generated by applying
constant
force in optical-tweezer experiments,
show that RNA hairpins and the P5abc subdomain
of the group I intron unfold reversibly. We use
coarse-grained Go-like models for RNA hairpins to explore
forced unfolding over a broad range of
temperatures. A number of predictions 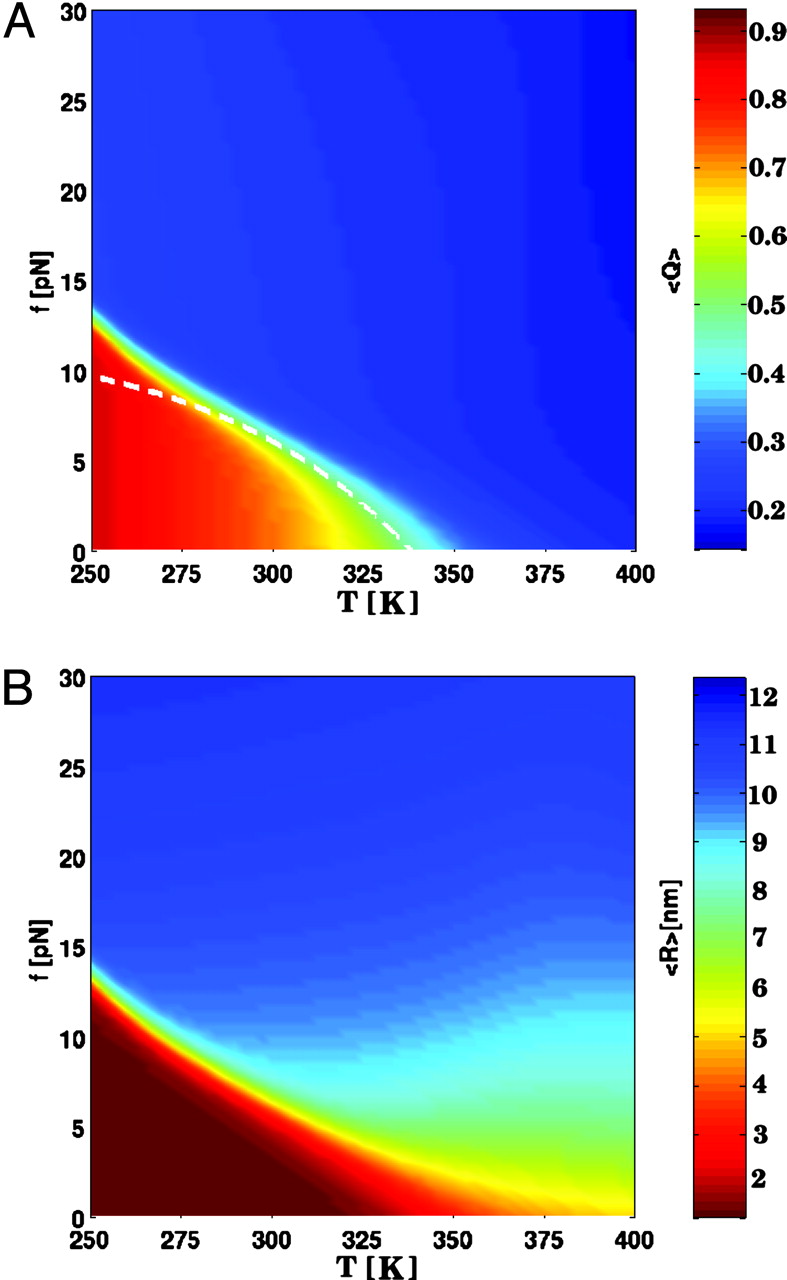 that
are amenable to experimental tests are made. At the
critical force, the hairpin jumps between folded and
unfolded conformations without populating
any discernible intermediates. The phase diagram in the
force vs temperature (f, T) plane shows that
the hairpin unfolds by an all-or-none
process. The cooperativity of the unfolding transition
increases dramatically at low temperatures.
Free energy of stability, obtained from time
averages of mechanical unfolding trajectories, coincides with
ensemble averages, which establishes ergodicity.The hopping time
between the native basin of attraction (NBA) and the unfolded
basin increases dr that
are amenable to experimental tests are made. At the
critical force, the hairpin jumps between folded and
unfolded conformations without populating
any discernible intermediates. The phase diagram in the
force vs temperature (f, T) plane shows that
the hairpin unfolds by an all-or-none
process. The cooperativity of the unfolding transition
increases dramatically at low temperatures.
Free energy of stability, obtained from time
averages of mechanical unfolding trajectories, coincides with
ensemble averages, which establishes ergodicity.The hopping time
between the native basin of attraction (NBA) and the unfolded
basin increases dr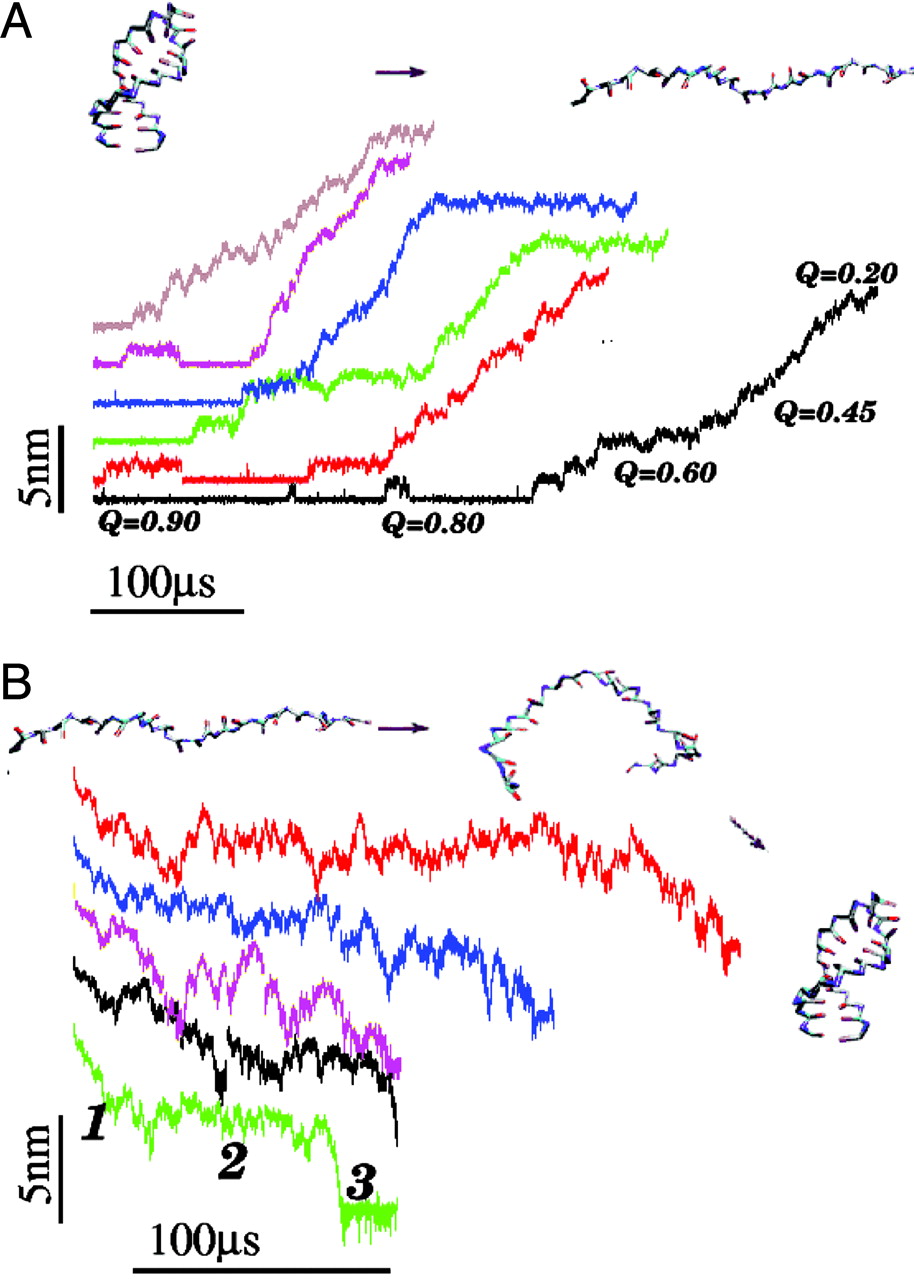 amatically along the phase
boundary. Thermal unfolding is stochastic,
whereas mechanical unfolding occurs in
"quantized steps" with great variations in the step
lengths. Refolding times, upon force quench, from stretched
states to the NBA are at least an order of magnitude
greater than folding times by temperature quench. Upon
force quench from stretched states, the NBA is reached in
at least three stages. In the initial stages, the mean
end-to-end distance decreases nearly continuously, and
there is a sudden transition to the NBA only in the
last stage. Because of the generality of the results, we
propose that similar behavior should be observed in
force quench refolding of proteins. amatically along the phase
boundary. Thermal unfolding is stochastic,
whereas mechanical unfolding occurs in
"quantized steps" with great variations in the step
lengths. Refolding times, upon force quench, from stretched
states to the NBA are at least an order of magnitude
greater than folding times by temperature quench. Upon
force quench from stretched states, the NBA is reached in
at least three stages. In the initial stages, the mean
end-to-end distance decreases nearly continuously, and
there is a sudden transition to the NBA only in the
last stage. Because of the generality of the results, we
propose that similar behavior should be observed in
force quench refolding of proteins.
Mechanical unfolding of RNA: from Hairpins to structures with internal
multiloops (Biophys. J. '07)
Mechanical
unfolding of RNA structures, ranging from hairpins to
ribozymes, using laser optical tweezer experiments have begun to
reveal the features of the energy landscape that cannot be easily
explored using conventional experiments. Upon application of
constant force (f), RNA hairpins undergo cooperative transitions
from folded to unfolded states whereas
subdomains of ribozymes unravel one at a time. Here, we use
a self-organized polymer model and Brownian dynamics
simulations to probe mechanical unfolding at constant force
and constant-loading rate of four RNA structures of varying
complexity. For simple hairpins, such as P5GA, application
of constant force or constant loading rate results in
bistable cooperative transitions between folded and unfolded
states without populating any intermediates. The transition state
location (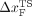 ) changes
dramatically as the loading rate is varied. At loading rates comparable
to those used in laser optical tweezer experiments, the
hairpin is plastic, with ) changes
dramatically as the loading rate is varied. At loading rates comparable
to those used in laser optical tweezer experiments, the
hairpin is plastic, with 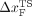 being midway between folded and unfolded states; whereas
at high loading rates, being midway between folded and unfolded states; whereas
at high loading rates, 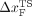 moves close to the folded state, i.e., RNA is brittle. For
the 29-nucleotide TAR RNA with the three-nucleotide bulge,
unfolding occurs in a nearly two-state manner with an
occasional pause in a high free energy metastable state.
Forced unfolding of the 55 nucleotides of the Hepatitis
IRES domain IIa, which has a distorted L-shaped structure,
results in well-populated stable intermediates. The most
stable force-stabilized intermediate represents straightening of
the L-shaped structure. For these structures, the unfolding pathways
can be predicted using the contact map of the native structures.
Unfolding of a RNA motif with internal multiloop, namely,
the 109-nucleotide prohead RNA that is part of the moves close to the folded state, i.e., RNA is brittle. For
the 29-nucleotide TAR RNA with the three-nucleotide bulge,
unfolding occurs in a nearly two-state manner with an
occasional pause in a high free energy metastable state.
Forced unfolding of the 55 nucleotides of the Hepatitis
IRES domain IIa, which has a distorted L-shaped structure,
results in well-populated stable intermediates. The most
stable force-stabilized intermediate represents straightening of
the L-shaped structure. For these structures, the unfolding pathways
can be predicted using the contact map of the native structures.
Unfolding of a RNA motif with internal multiloop, namely,
the 109-nucleotide prohead RNA that is part of the  29 DNA packaging
motor, at constant value of rf occurs with three
distinct rips that represent unraveling
of the paired helices. The rips represent kinetic barriers
to unfolding. Our work shows 1), the response of RNA to
force is largely determined by the native structure; and
2), only by probing mechanical unfolding over a wide range
of forces can the underlying energy landscape be fully
explored. 29 DNA packaging
motor, at constant value of rf occurs with three
distinct rips that represent unraveling
of the paired helices. The rips represent kinetic barriers
to unfolding. Our work shows 1), the response of RNA to
force is largely determined by the native structure; and
2), only by probing mechanical unfolding over a wide range
of forces can the underlying energy landscape be fully
explored.
Pathways and
Kinetic Barriers in Mechanical Unfolding and Refolding of RNA and
Proteins (Structure '06)
Using
self-organized polymer models, we predict mechanical unfolding
and refolding pathways of ribozymes, and the green fluorescent protein.
In agreement with experiments, there are between six and eight
unfolding transitions in the Tetrahymena ribozyme. Depending on
the loading rate, the number of rips in the force-ramp unfolding of the
Azoarcus ribozymes is between two and
four. Force-quench refolding of the P4-P6 subdomain of the Tetrahymena
ribozyme occurs through a compact intermediate. Subsequent formation of
tertiary contacts between helices P5b-P6a and P5a/P5c-P4 leads to the
native state. The force-quench refolding pathways agree with ensemble
experiments. In the dominant unfolding route, the N-terminal  helix of GFP unravels first, followed by disruption of the N terminus
占쏙옙
strand. There is a third intermediate that involves disruption of three
other strands. In accord with experiments, the force-quench refolding
pathway of GFP is hierarchic, with the rate-limiting step being the
closure of the barrel.
helix of GFP unravels first, followed by disruption of the N terminus
占쏙옙
strand. There is a third intermediate that involves disruption of three
other strands. In accord with experiments, the force-quench refolding
pathway of GFP is hierarchic, with the rate-limiting step being the
closure of the barrel.

|
|
|
Contact Information : Changbong Hyeon,
Professor, School of Computational Sciences, Korea Institute for Advanced Study
, Seoul 02455, Republic of Korea
+82-2-958-3810 (tel)
|
|
© 2010 KIAS Theoretical and Computational Biophysics
Group |
|
|